Introduction
In this tutorial, we will see how to train a custom YOLO model and use it with PyFastTrack to track objects.
There are three parts to this tutorial:
- Annotate the dataset.
- Train the model.
- Use the model.
This tutorial will not require GPU; it will use Google Colaboratory to train the model.
Model annotation
PyFastTrack requires a YOLO detector that performs segmentation, i.e., that can find which pixels belong to the object and which pixels are not.
Training the model is the most tedious task. There are strategies to reduce the tediousness, but we will focus on the basic approach. We will use LabelMe to annotate manually.
The goal is to draw a polygon to delimitate the object (or objects) and assign it to its class. Several exportation formats are possible in LabelMe, and here, we use the default JSON format that we later convert to generate a formatted YOLO dataset.
Train the model
First, we must convert the dataset to a format compatible with YOLO. We use labelme2yolo tool:
!pip install labelme2yolo
python -m labelme2yolo --json_dir /path/to/labelme_json_dir/ --val_size 0 --test_size 0.15
That produces a dataset with an architecture as follows.
/path/to/labelme_json_dir/
├─ YOLODataset/
├─ dataset.yaml
├─ labels/
│ ├─ train/
│ ├─ test/
├─ images/
│ ├─ train/
│ ├─ test/
For segmentation, each image is associated with a text label file. It contains the class number and the polygon coordinates to create the segmentation mask, one line per object. The coordinates must be normalized between 0 and 1 (x divided by the image width and y divided by the image height).
Training deep learning models without GPU can be time-consuming or even impossible. Alternatives exist by renting a GPU instance from a provider (OVH, AWS, Google, etc.). A more straightforward option to tinker with a small model is Google Colaboratory.
Using Google Colaboratory is relatively straightforward. The first step is to upload the dataset folder to your Google Drive. The second step is to create a Colab notebook, set a GPU for processing using Edit>Notebook settings, and choose GPU.
Inside the notebook, we first mount the Google Drive:
import os
from google.colab import drive
drive.mount('/content/drive')
os.chdir('/content/drive/MyDrive/model/')
Then install the dependencies for YOLOv8:
!pip install ultralytics
To train the model, we use the command below (complete documentation can be found here):
from ultralytics import YOLO
model = YOLO('yolov8m-seg.yaml')
model.train(data='YOLODataset/dataset.yaml', batch=8, epochs=200, imgsz=640)
Google Colaboratory enforces a limitation on GPU usage and runtime time. You can circumvent these limitations by training in several steps adding --resume to the training command to continue the training, or by subscribing to a Colaboratory premium. The resulting weights can be downloaded from Google Drive (YOLODataset/runs/segment) and used for inference.
Use the model
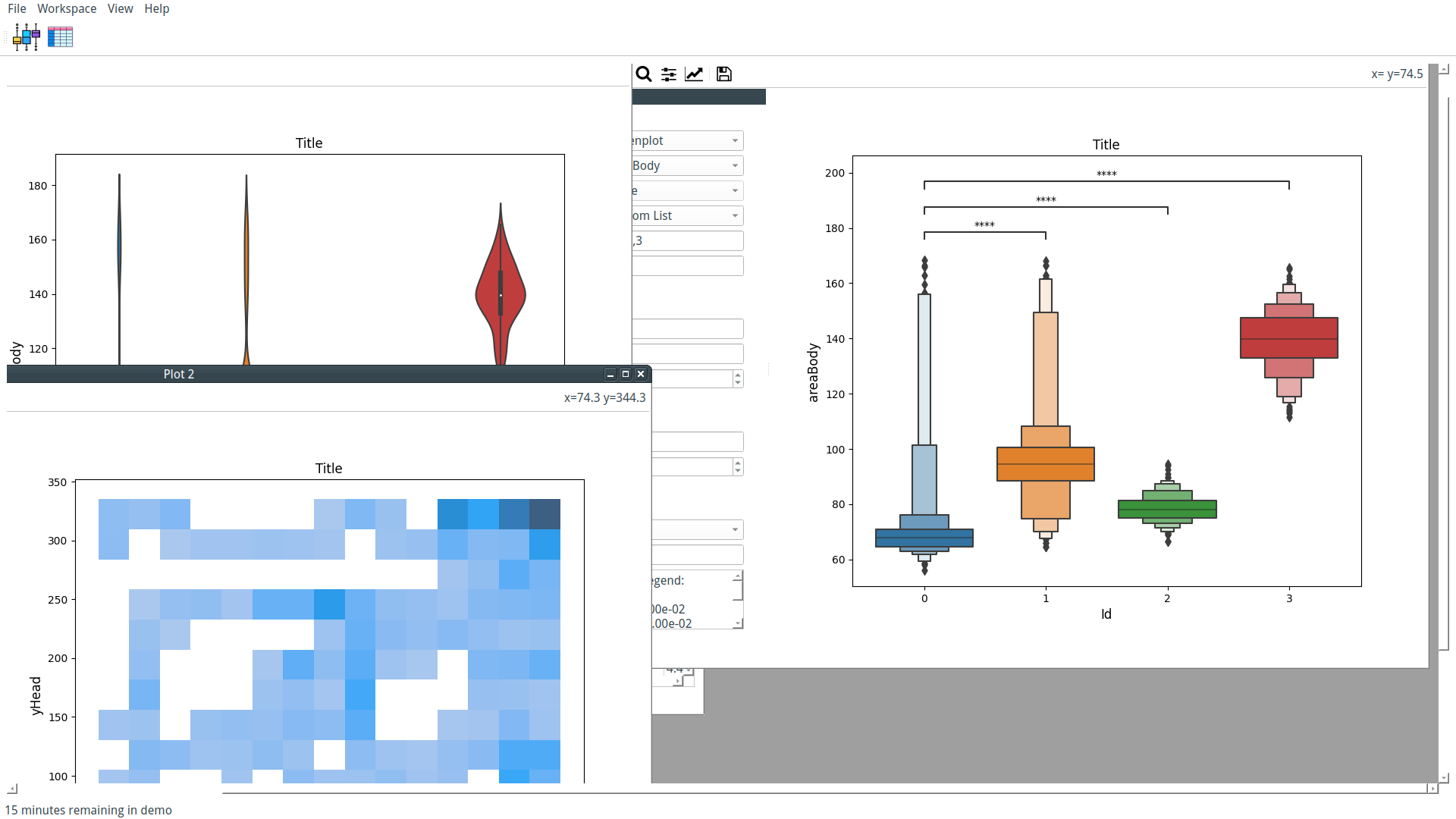
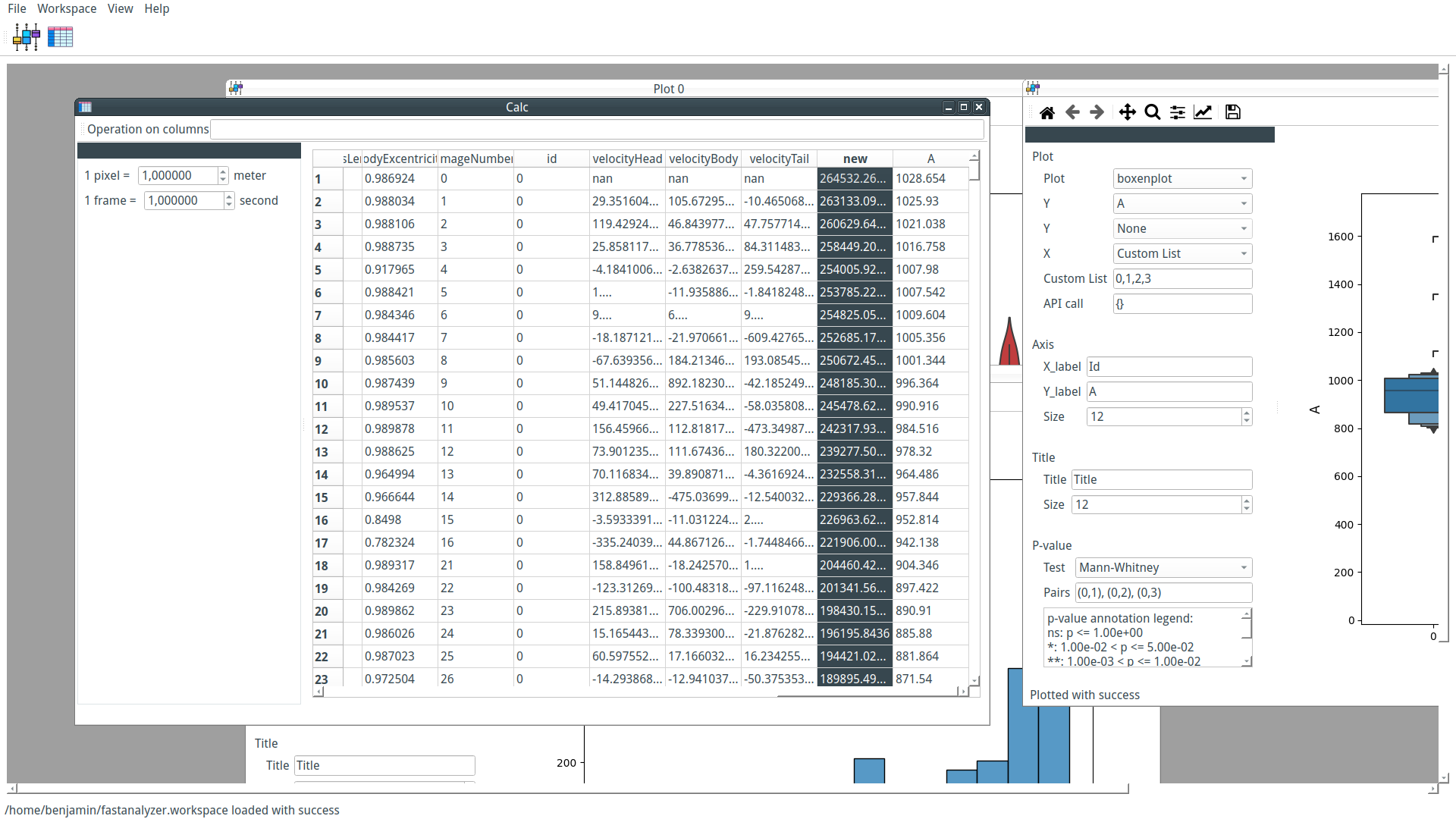
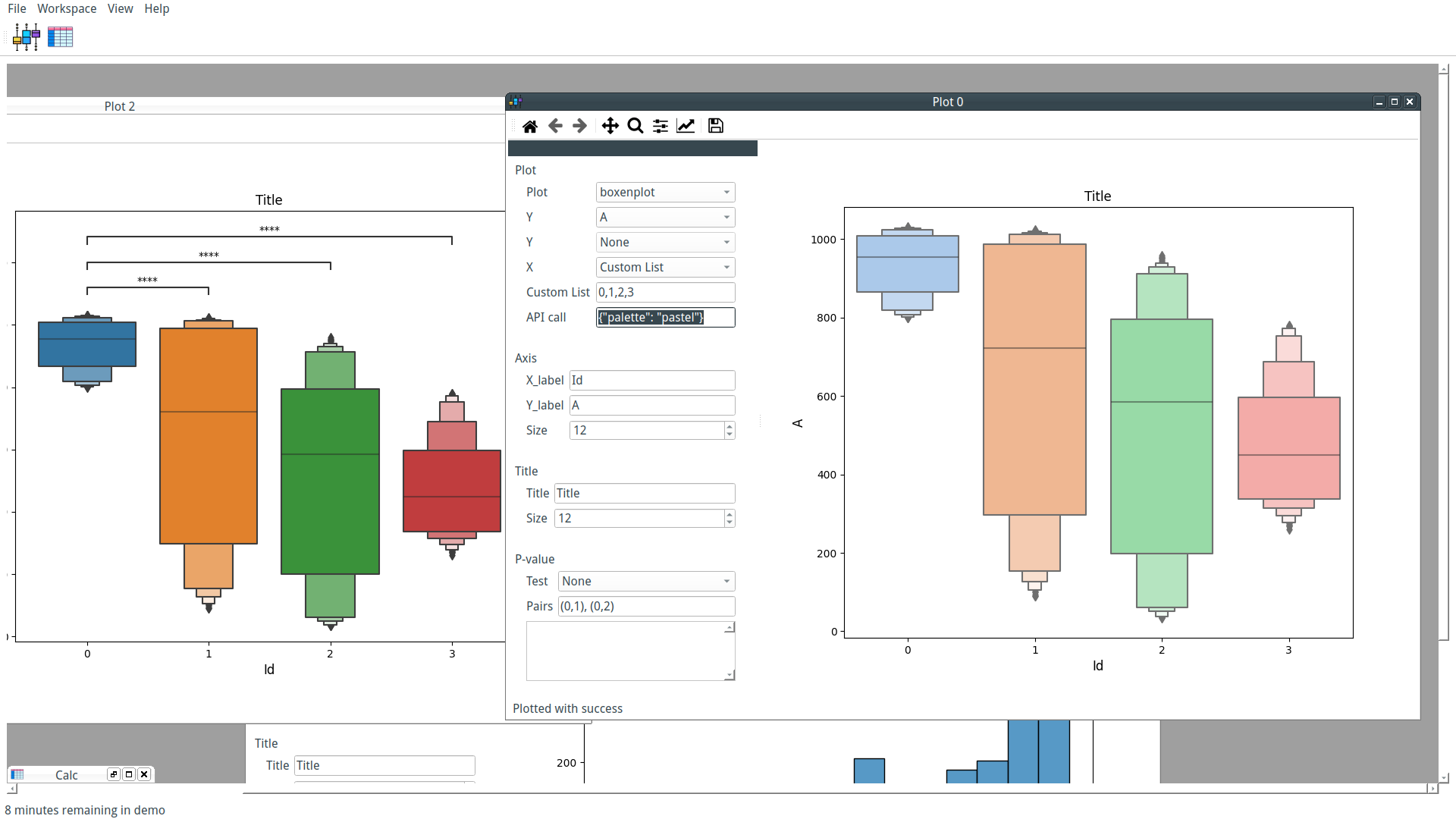
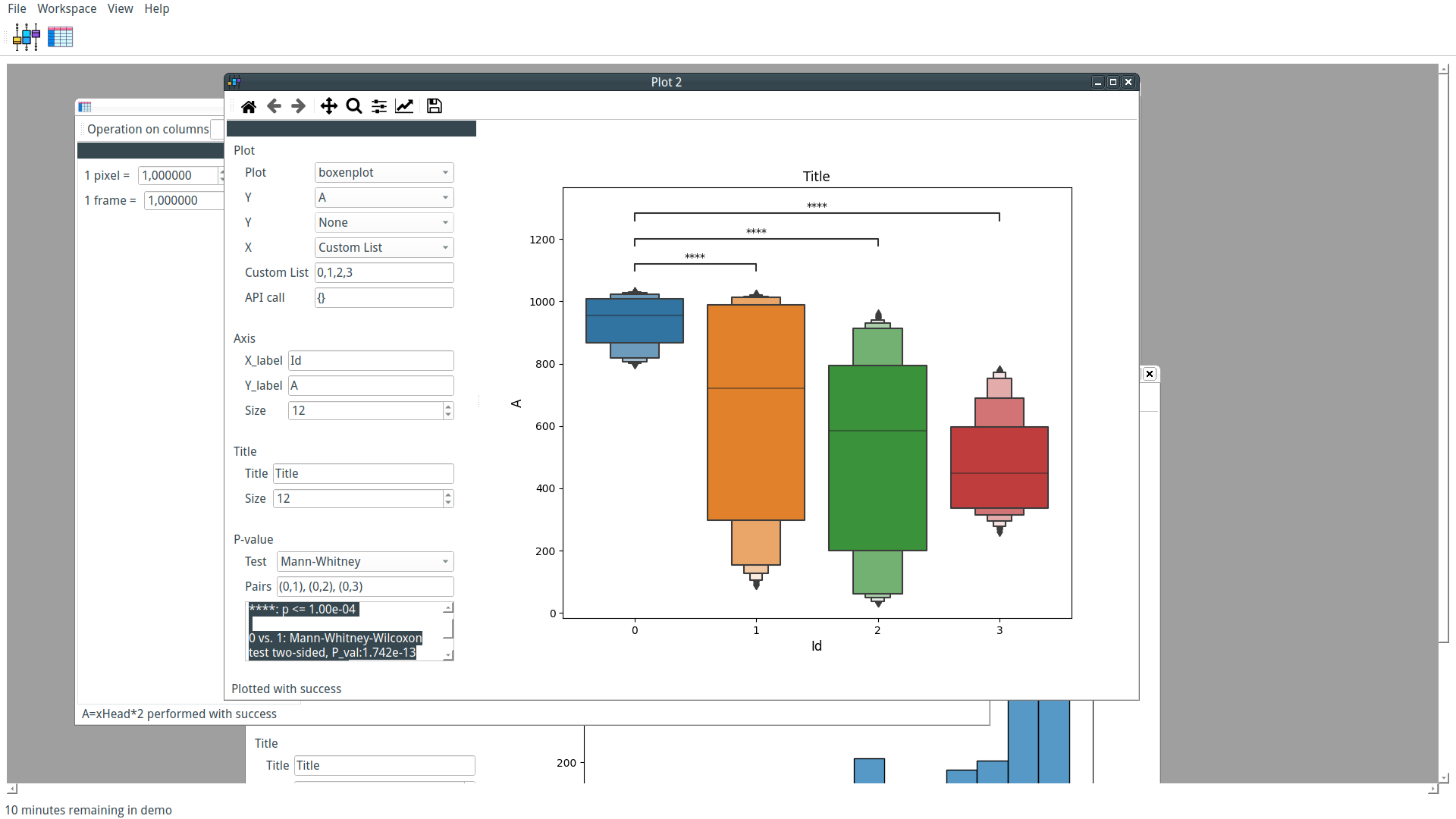
Our custom-trained model can now be used by PyFastTrack to perform the tracking (the tracking can also be done in Google Collab if you want to leverage PyFastTrack GPU capability). The procedure to follow is detailed in the PyFastTrack documentation, and it is straightforward:
- Setup the detector
- Setup the tracker
- Track and save/use tracking data
!pip install pyfasttrack
from pyfasttrack.yolo_detector import YoloDetector
from pyfasttrack.tracker import Tracker
from pyfasttrack.data import Result
import cv2
import os
# Data saver
saver = Result("test/data/images/")
# Set up detector
# See https://github.com/ultralytics/ultralytics/blob/44c7c3514d87a5e05cfb14dba5a3eeb6eb860e70/ultralytics/datasets/coco.yaml for equivalence between coco labels and indexes
yolo_params = {"model": "my_model.pt", "conf": 0.5}
detector = YoloDetector(yolo_params)
# Set up tracker
params = {"spot": 2, "normDist": 1, "normAngle": 2,
"normArea": 1, "normPerim": 1, "maxDist": 500, "maxTime": 100}
tracker = Tracker()
tracker.set_params(params)
tracker.set_detector(detector)
camera = cv2.VideoCapture(
"/test/data/images/video.mp4")
dat = tracker.initialize(camera.read()[1])
saver.add_data(dat)
ret = True
while (ret):
ret, frame = camera.read()
if ret:
dat = tracker.process(frame)
saver.add_data(dat)
camera.release()